Part 1: Neurons and simple neural networks
Introduction
In this handout we cover the first steps in using PyNEST to simulate neuronal networks. When you have worked through this material, you will know how to:
- start PyNEST
- create neurons and stimulating/recording devices
- query and set their parameters
- connect them to each other or to devices
- simulate the network
- extract the data from recording devices
For more information on the usage of PyNEST, please see the other sections of this primer:
- Part 2: Populations of neurons
- Part 3: Connecting networks with synapses
- Part 4: Topologically structured networks
More advanced examples can be found at Example Networks, or have a look at at the source directory of your NEST installation in the subdirectory: pynest/examples/.
PyNEST - an interface to the NEST simulator
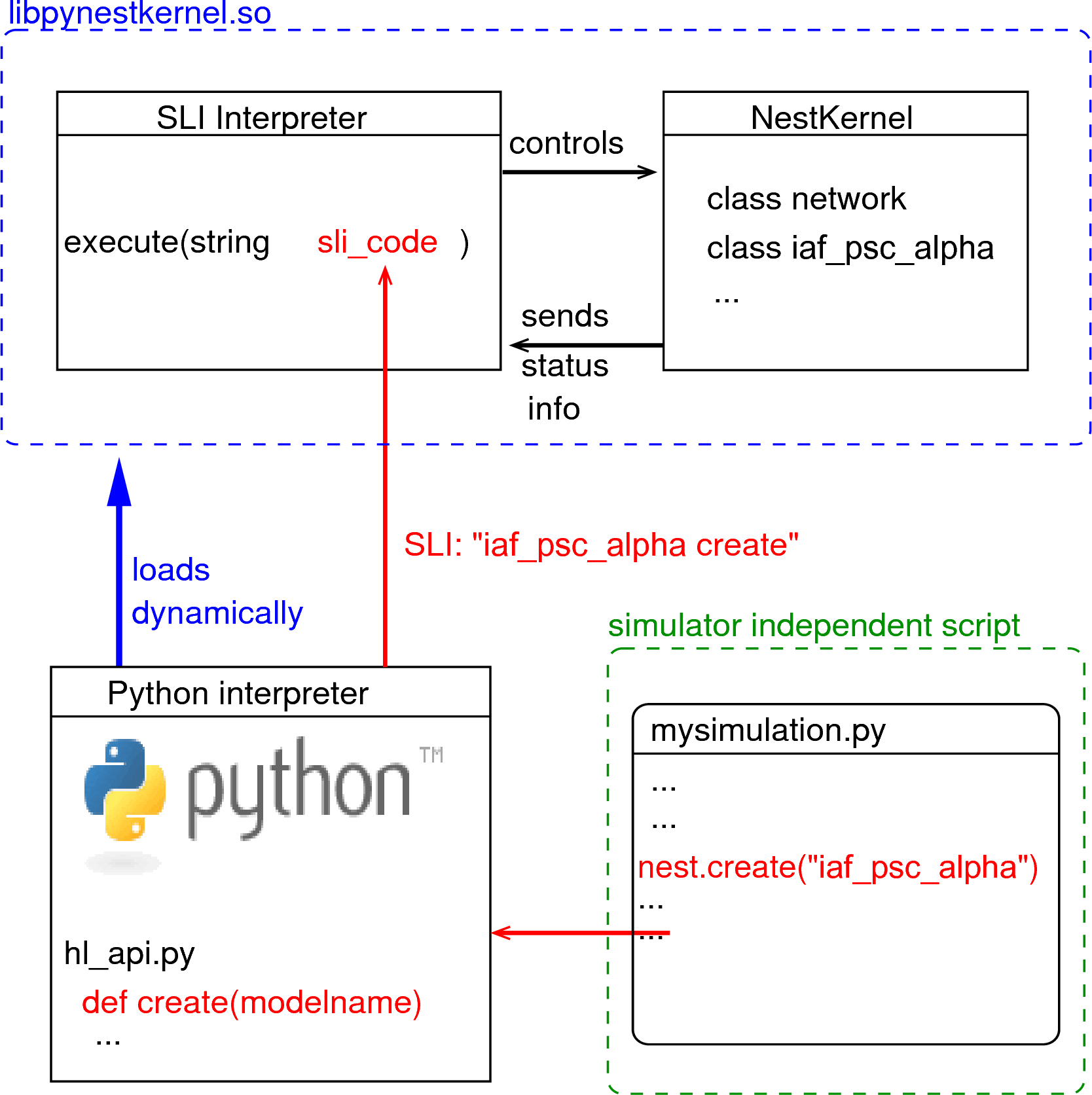
|
Figure 1: The Python interpreter imports NEST as a module and dynamically loads the NEST simulator kernel (pynestkernel.so). The core functionality is defined in hl_api.py. A simulation script of the user (mysimulation.py) uses functions defined in this high-level API. These functions generate code in SLI (Simulation Language Interpreter), the native language of the interpreter of NEST. This interpreter, in turn, controls the NEST simulation kernel.
The NEural Simulation Tool (NEST: www.nest-initiative.org) is designed for the simulation of large heterogeneous networks of point neurons. It is open source software released under the GPL licence. The simulator comes with an interface to Python [4]. Fig. 1 illustrates the interaction between the user’s simulation script (mysimulation.py) and the NEST simulator. [2] contains a technically detailed description of the implementation of this interface and parts of this text are based on this reference. The simulation kernel is written in C++ to obtain highest possible performance for the simulation.
You can use PyNEST interactively from the Python prompt or from within ipython. This is very helpful when you are exploring PyNEST, trying to learn a new functionality or debugging a routine. Once out of the exploratory mode, you will find it saves a lot of time to write your simulations in text files. These can in turn be run from the command line or from the Python or ipython prompt.
Whether working interactively, semi-interactively, or purely executing scripts, the first thing that needs to happen is importing NEST’s functionality into the Python interpreter.
import nestIt should be noted, however, that certain external packages must be imported before importing nest. These include sklearn (scikit-learn) and scipy (SciPy):
from sklearn.svm import LinearSVC
from scipy.special import erf
import nestAs with every other module for Python, the available functions can be prompted for.
dir(nest)One such command is nest.Models?, which will return a list of all the available models you can use. If you want to obtain more information about a particular command, you may use IPython’s standard help system.
nest.Models?This will return the help text (docstring) explaining the use of this particular function. There is a help system within NEST as well. You can open the help pages in a browser using nest.helpdesk() and you can get the help page for a particular object using nest.help(object).
Creating Nodes
A neural network in NEST consists of two basic element types: nodes and connections. Nodes are either neurons, devices or sub-networks. Devices are used to stimulate neurons or to record from them. Nodes can be arranged in sub-networks to build hierarchical networks such as layers, columns, and areas - we will get to this later in the course. For now we will work in the default sub-network which is present when we start NEST, known as the root node.
To begin with, the root sub-network is empty. New nodes are created with the command Create, which takes as arguments the model name of the desired node type, and optionally the number of nodes to be created and the initialising parameters. The function returns a list of handles to the new nodes, which you can assign to a variable for later use. These handles are integer numbers, called ids. Many PyNEST functions expect or return a list of ids (see Sec.8). Thus it is easy to apply functions to large sets of nodes with a single function call.
After having imported NEST and also the Pylab interface to Matplotlib [[3]] (#3), which we will use to display the results, we can start creating nodes. As a first example, we will create a neuron of type iaf_psc_alpha. This neuron is an integrate-and-fire neuron with alpha-shaped postsynaptic currents. The function returns a list of the ids of all the created neurons, in this case only one, which we store in a variable called neuron.
import pylab
import nest
neuron = nest.Create("iaf_psc_alpha")We can now use the id to access the properties of this neuron. Properties of nodes in NEST are generally accessed via Python dictionaries of key-value pairs of the form {key: value}. In order to see which properties a neuron has, you may ask it for its status.
nest.GetStatus(neuron)This will print out the corresponding dictionary in the Python console. Many of these properties are not relevant for the dynamics of the neuron. To find out what the interesting properties are, look at the documentation of the model through the helpdesk. If you already know which properties you are interested in, you can specify a key, or a list of keys, as an optional argument to GetStatus:
nest.GetStatus(neuron, "I_e")
nest.GetStatus(neuron, ["V_reset", "V_th"])In the first case we query the value of the constant background current I_e; the result is given as a tuple with one element. In the second case, we query the values of the reset potential and threshold of the neuron, and receive the result as a nested tuple. If GetStatus is called for a list of nodes, the dimension of the outer tuple is the length of the node list, and the dimension of the inner tuples is the number of keys specified.
To modify the properties in the dictionary, we use SetStatus. In the following example, the background current is set to 376.0pA, a value causing the neuron to spike periodically.
nest.SetStatus(neuron, {"I_e": 376.0})Note that we can set several properties at the same time by giving multiple comma separated key:value pairs in the dictionary. Also be aware that NEST is type sensitive - if a particular property is of type double, then you do need to explicitly write the decimal point:
nest.SetStatus(neuron, {"I_e": 376})will result in an error. This conveniently protects us from making integer division errors, which are hard to catch.
Next we create a multimeter, a device we can use to record the membrane voltage of a neuron over time. We set its property withtime such that it will also record the points in time at which it samples the membrane voltage. The property record_from expects a list of the names of the variables we would like to record. The variables exposed to the multimeter vary from model to model. For a specific model, you can check the names of the exposed variables by looking at the neuron’s property recordables.
multimeter = nest.Create("multimeter")
nest.SetStatus(multimeter, {"withtime":True, "record_from":["V_m"]})We now create a spikedetector, another device that records the spiking events produced by a neuron. We use the optional keyword argument params to set its properties. This is an alternative to using SetStatus. The property withgid indicates whether the spike detector is to record the source id from which it received the event (i.e. the id of our neuron).
spikedetector = nest.Create("spike_detector",
params={"withgid": True, "withtime": True})A short note on naming: here we have called the neuron neuron, the multimeter multimeter and so on. Of course, you can assign your created nodes to any variable names you like, but the script is easier to read if you choose names that reflect the concepts in your simulation.
Connecting nodes with default connections
Now we know how to create individual nodes, we can start connecting them to form a small network.
nest.Connect(multimeter, neuron)
nest.Connect(neuron, spikedetector)
A 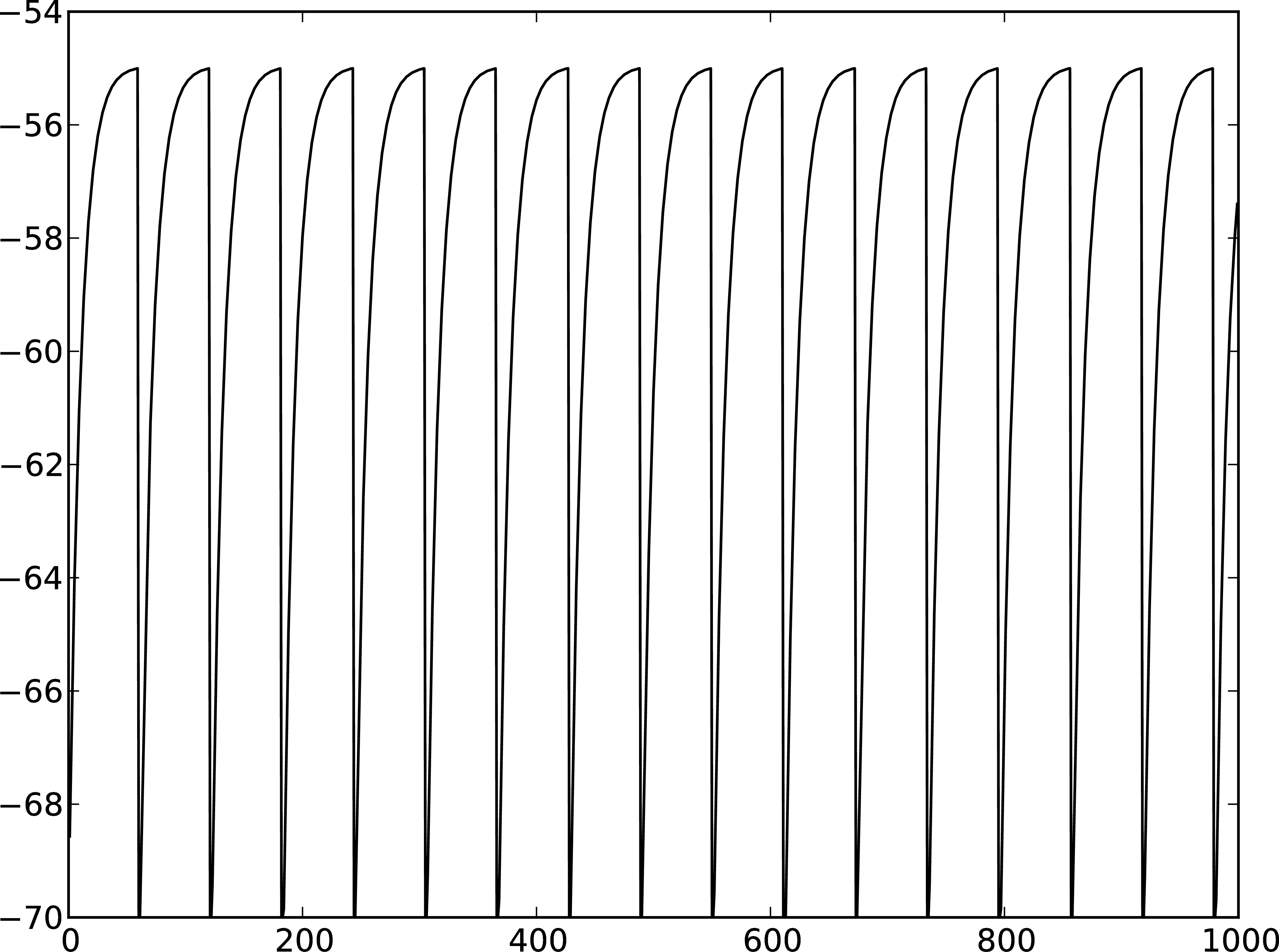
|
B 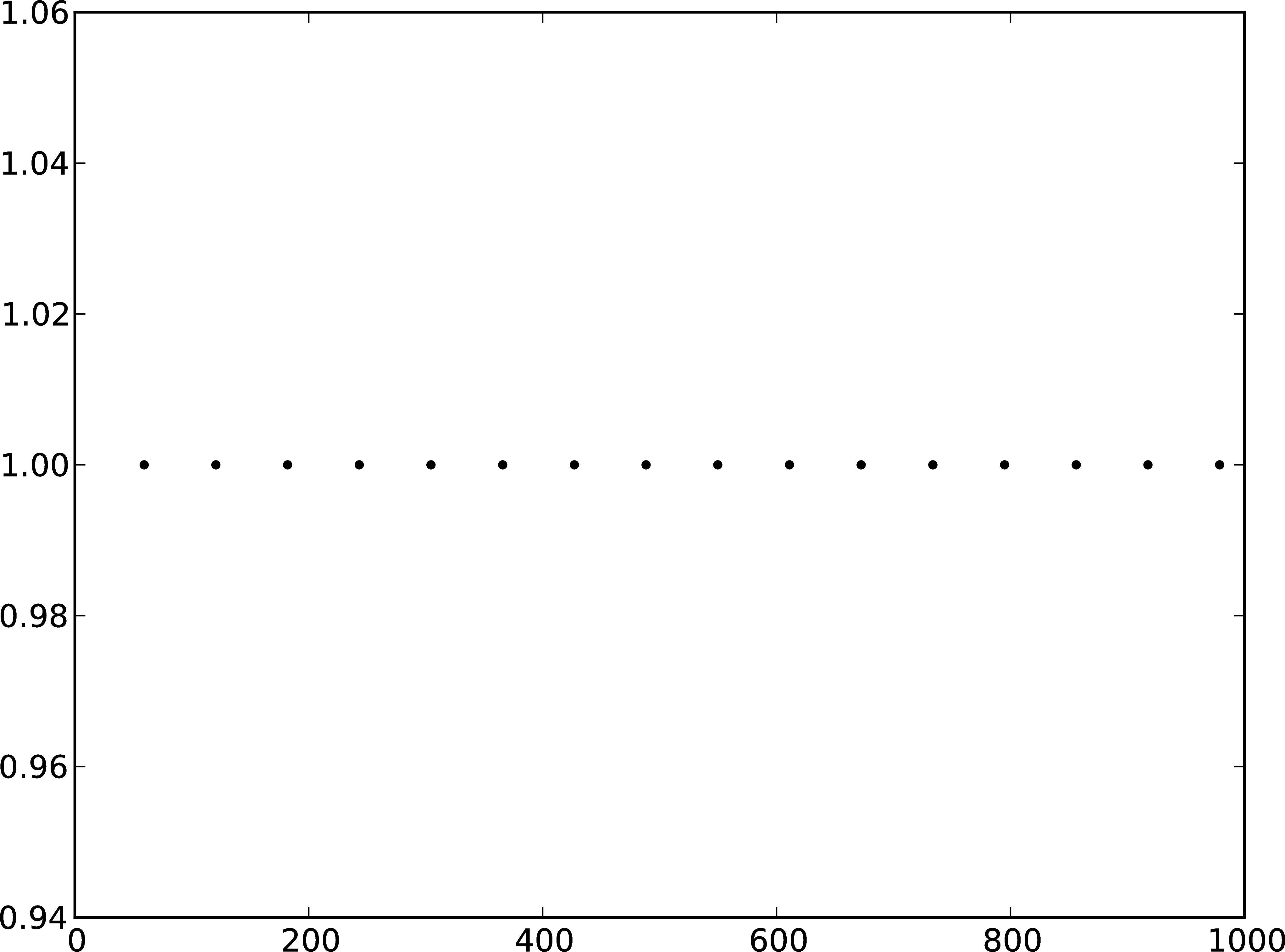
|
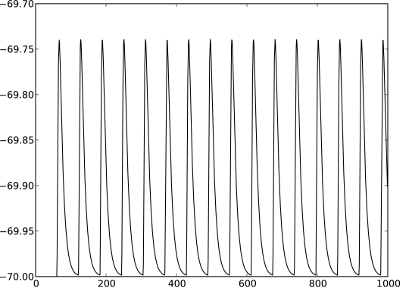
Figure 2: A Membrane potential of integrate-and-fire neuron with constant input current. B Spikes of the neuron.
The order in which the arguments to Connect are specified reflects the flow of events: if the neuron spikes, it sends an event to the spike detector. Conversely, the multimeter periodically sends requests to the neuron to ask for its membrane potential at that point in time. This can be regarded as a perfect electrode stuck into the neuron.
Now we have connected the network, we can start the simulation. We have to inform the simulation kernel how long the simulation is to run. Here we choose 1000ms.
nest.Simulate(1000.0)Congratulations, you have just simulated your first network in NEST!
Extracting and plotting data from devices
After the simulation has finished, we can obtain the data recorded by the multimeter.
dmm = nest.GetStatus(multimeter)[0]
Vms = dmm["events"]["V_m"]
ts = dmm["events"]["times"]In the first line, we obtain the list of status dictionaries for all queried nodes. Here, the variable multimeter is the id of only one node, so the returned list just contains one dictionary. We extract the first element of this list by indexing it (hence the [0] at the end). This type of operation occurs quite frequently when using PyNEST, as most functions are designed to take in and return lists, rather than individual values. This is to make operations on groups of items (the usual case when setting up neuronal network simulations) more convenient.
This dictionary contains an entry named events which holds the recorded data. It is itself a dictionary with the entries V_m and times, which we store separately in Vms and ts, in the second and third line, respectively. If you are having trouble imagining dictionaries of dictionaries and what you are extracting from where, try first just printing dmm to the screen to give you a better understanding of its structure, and then in the next step extract the dictionary events, and so on.
Now we are ready to display the data in a figure. To this end, we make use of pylab.
import pylab
pylab.figure(1)
pylab.plot(ts, Vms)The second line opens a figure (with the number 1), and the third line actually produces the plot. You can’t see it yet because we have not used pylab.show(). Before we do that, we proceed analogously to obtain and display the spikes from the spike detector.
dSD = nest.GetStatus(spikedetector,keys="events")[0]
evs = dSD["senders"]
ts = dSD["times"]
pylab.figure(2)
pylab.plot(ts, evs, ".")
pylab.show()Here we extract the events more concisely by using the optional keyword argument keys to GetStatus. This extracts the dictionary element with the key events rather than the whole status dictionary. The output should look like Fig. 2. If you want to execute this as a script, just paste all lines into a text file named, say, one-neuron.py . You can then run it from the command line by prefixing the file name with python, or from the Python or ipython prompt, by prefixing it with run.
It is possible to collect information of multiple neurons on a single multimeter. This does complicate retrieving the information: the data for each of the n neurons will be stored and returned in an interleaved fashion. Luckily Python provides us with a handy array operation to split the data easily: array slicing with a step (sometimes called stride). To explain this you have to adapt the model created in the previous part. Save your code under a new name, in the next section you will also work on this code. Create an extra neuron with the background current given a different value:
neuron2 = nest.Create("iaf_psc_alpha")
nest.SetStatus(neuron2 , {"I_e": 370.0})now connect this newly created neuron to the multimeter:
nest.Connect(multimeter, neuron2)Run the simulation and plot the results, they will look incorrect. To fix this you must plot the two neuron traces separately. Replace the code that extracts the events from the multimeter with the following lines.
pylab.figure(2)
Vms1 = dmm["events"]["V_m"][::2] # start at index 0: till the end: each second entry
ts1 = dmm["events"]["times"][::2]
pylab.plot(ts1, Vms1)
Vms2 = dmm["events"]["V_m"][1::2] # start at index 1: till the end: each second entry
ts2 = dmm["events"]["times"][1::2]
pylab.plot(ts2, Vms2)Additional information can be found at <http://docs.scipy.org/doc/numpy-1.10 .0/reference/arrays.indexing.html>.
Connecting nodes with specific connections
A commonly used model of neural activity is the Poisson process. We now adapt the previous example so that the neuron receives 2 Poisson spike trains, one excitatory and the other inhibitory. Hence, we need a new device, the poisson_generator. After creating the neurons, we create these two generators and set their rates to 80000Hz and 15000Hz, respectively.
noise_ex = nest.Create("poisson_generator")
noise_in = nest.Create("poisson_generator")
nest.SetStatus(noise_ex, {"rate": 80000.0})
nest.SetStatus(noise_in, {"rate": 15000.0})Additionally, the constant input current should be set to 0:
nest.SetStatus(neuron, {"I_e": 0.0})Each event of the excitatory generator should produce a postsynaptic current of 1.2pA amplitude, an inhibitory event of -2.0pA. The synaptic weights can be defined in a dictionary, which is passed to the Connect function using the keyword syn_spec (synapse specifications). In general all parameters determining the synapse can be specified in the synapse dictionary, such as "weight", "delay", the synaptic model ("model") and parameters specific to the synaptic model.
syn_dict_ex = {"weight": 1.2}
syn_dict_in = {"weight": -2.0}
nest.Connect([noise_ex], neuron, syn_spec=syn_dict_ex)
nest.Connect([noise_in], neuron, syn_spec=syn_dict_in)
A 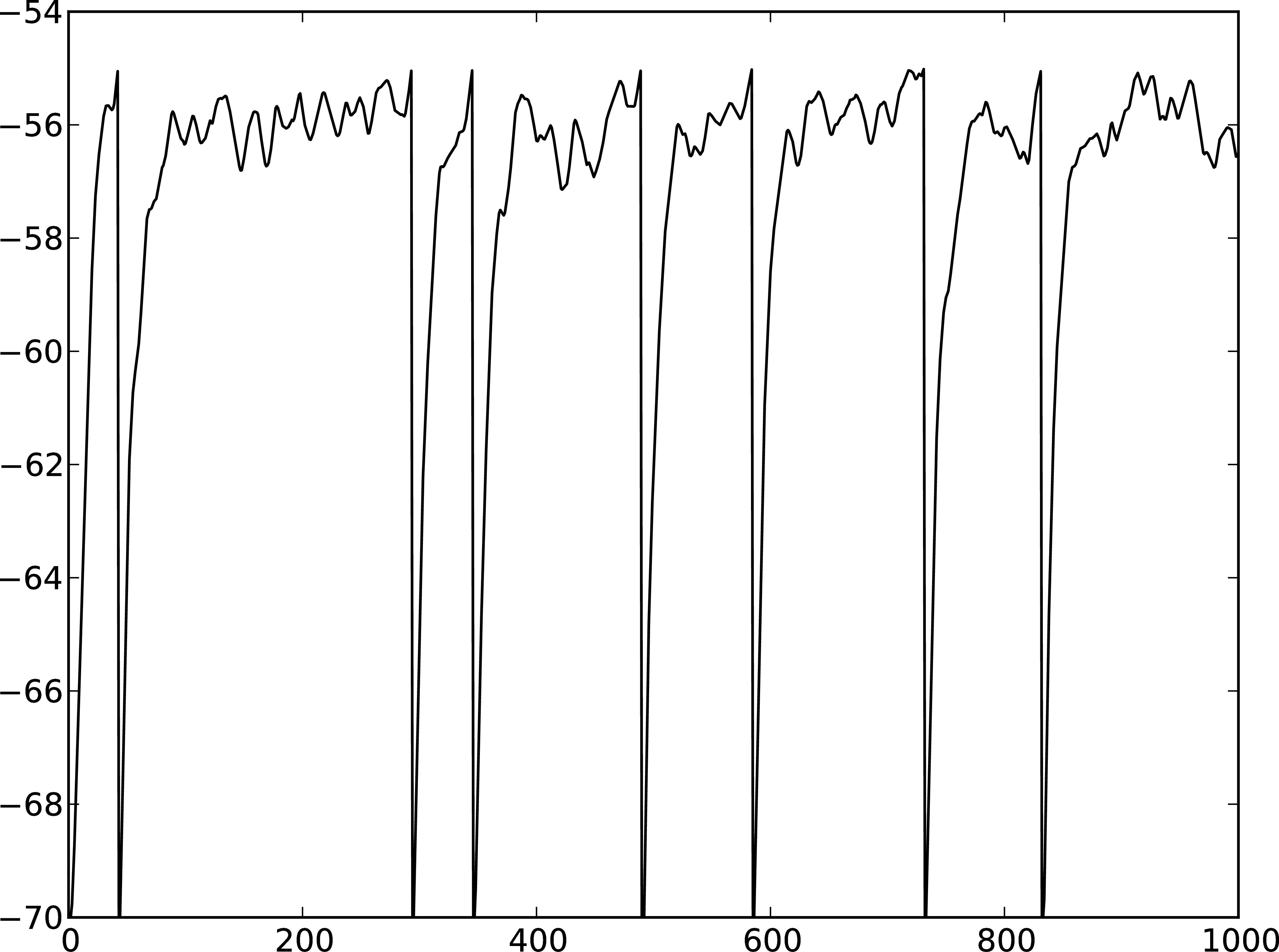
|
B 
|
Figure 3: A Membrane potential of integrate-and-fire neuron with Poisson noise as input. B Spikes of the neuron.
The rest of the code remains as before. You should see a membrane potential as in Fig. 3.
In the next part of the introduction (Part 2: Populations of neurons) we will look at more methods for connecting many neurons at once.
Two connected neurons
|
Figure 4: Postsynaptic potentials in neuron2 evoked by the spikes of neuron1
There is no additional magic involved in connecting neurons. To demonstrate this, we start from our original example of one neuron with a constant input current, and add a second neuron.
import pylab
import nest
neuron1 = nest.Create("iaf_psc_alpha")
nest.SetStatus(neuron1, {"I_e": 376.0})
neuron2 = nest.Create("iaf_psc_alpha")
multimeter = nest.Create("multimeter")
nest.SetStatus(multimeter, {"withtime":True, "record_from":["V_m"]})We now connect neuron1 to neuron2, and record the membrane potential from neuron2 so we can observe the postsynaptic potentials caused by the spikes of neuron1.
nest.Connect(neuron1, neuron2, syn_spec = {"weight":20.0})
nest.Connect(multimeter, neuron2)Here the default delay of 1ms was used. If the delay is specified in addition to the weight, the following shortcut is available:
nest.Connect(neuron1, neuron2, syn_spec={"weight":20, "delay":1.0})If you simulate the network and plot the membrane potential as before, you should then see the postsynaptic potentials of neuron2 evoked by the spikes of neuron1 as in Fig. 4.
Command overview
These are the functions we introduced for the examples in this handout; the following sections of this introduction will add more.
Getting information about NEST
Models(mtype="all", sel=None):
Return a list of all available models (nodes and synapses). Usemtype="nodes"to only see node models,mtype="synapses"to only see synapse models.selcan be a string, used to filter the result list and only return models containing it.helpdesk(browser="firefox"):
Opens the NEST documentation pages in the given browser.help(obj=None,pager="less"):
Opens the help page for the given object.
Nodes
Create(model, n=1, params=None):
Createninstances of typemodelin the current sub-network. Parameters for the new nodes can be given asparams(a single dictionary, or a list of dictionaries with sizen). If omitted, themodel’s defaults are used.GetStatus(nodes, keys=None):
Return a list of parameter dictionaries for the given list ofnodes. Ifkeysis given, a list of values is returned instead.keysmay also be a list, in which case the returned list contains lists of values.SetStatus(nodes, params, val=None):
Set the parameters of the givennodestoparams, which may be a single dictionary, or a list of dictionaries of the same size asnodes. Ifvalis given,paramshas to be the name of a property, which is set tovalon thenodes.valcan be a single value, or a list of the same size asnodes.
Connections
This is an abbreviated version of the documentation for the Connect function, please see NEST’s online help for the full version and Connection Management for an introduction and worked examples.
Connect(pre, post, conn_spec=None, syn_spec=None, model=None):
Connect pre neurons to post neurons.Neurons in pre and post are connected using the specified connectivity ("one_to_one"by default) and synapse type ("static_synapse"by default). Details depend on the connectivity rule. Note: Connect does not iterate over subnets, it only connects explicitly specified nodes.pre- presynaptic neurons, given as list of GIDspost- presynaptic neurons, given as list of GIDsconn_spec- name or dictionary specifying connectivity rule, see belowsyn_spec- name or dictionary specifying synapses, see below
Connectivity
Connectivity is either specified as a string containing the name of a connectivity rule (default: "one_to_one") or as a dictionary specifying the rule and rule-specific parameters (e.g. "indegree"), which must be given. In addition switches allowing self-connections ("autapses", default: True) and multiple connections between a pair of neurons ("multapses", default: True) can be contained in the dictionary.
Synapse
The synapse model and its properties can be inserted either as a string describing one synapse model (synapse models are listed in the synapsedict) or as a dictionary as described below. If no synapse model is specified the default model "static_synapse" will be used. Available keys in the synapse dictionary are "model", "weight", "delay", "receptor_type" and parameters specific to the chosen synapse model. All parameters are optional and if not specified will use the default values determined by the current synapse model. "model" determines the synapse type, taken from pre-defined synapse types in NEST or manually specified synapses created via CopyModel(). All other parameters can be scalars or distributions. In the case of scalar parameters, all keys take doubles except for "receptor_type" which has to be initialised with an integer. Distributed parameters are initialised with yet another dictionary specifying the distribution ("distribution", such as "normal") and distribution-specific paramters (such as "mu" and "sigma").
Simulation control
Simulate(t):
Simulate the network fortmilliseconds.
References
[1] Marc-Oliver Gewaltig and Markus Diesmann. (NEural Simulation Tool). , 2(4):1430, 2007.
[2] J. M. Eppler, M. Helias, E. Muller, M. Diesmann, and M. Gewaltig. : a convenient interface to the NEST simulator. , 2:12, 2009.
[3] John D. Hunter. Matplotlib: A 2d graphics environment., 9(3):90–95, 2007.
[4] Python Software Foundation. The Python programming language, 2008. http://www.python.org.
